Advancing Informatics for Microbiology – Further Information
Throughout the long history of industrial and academic research, many microbes have been isolated, characterized and preserved (whenever possible) in culture collections. With the steady accumulation in observational data of biodiversity as well as microbial sequencing data, bio-resource centers have to function as data and information repositories to serve academia, industry, and regulators on behalf of and for the general public. Hence, the World Data Centre for Microorganisms (WDCM) started to take its responsibility for constructing an effective information environment that would promote and sustain microbial research data activities, and bridge the gaps currently present within and outside the microbiology communities.

Increasing demands on culture collections for authenticated, reliable biological material and associated information were accompanied by the growth of biotechnology and basic science. The WFCC guidelines recommend that every collection publish an online or printed catalogue regularly, both to disseminate information about strains and to promote scientific and industrial usage of materials held in their collection. However, according to the available statistics, fewer than one-sixth of collections registered in CCINFO post their catalogue online and this greatly hinders the visibility and hence the accessibility of strains in these collections without public electronic catalogs.
To help the collections to share their information in a standard and easy way, the current TG-AIM constructed a data management system and a global catalogue to organize, make public, and explore the data resources of its member collections. This data management system, called the WFCC Global Catalogue of Microorganisms (GCM) is a scalable, reliable, dynamic and user-friendly system that helps culture collections manage, disseminate and share the information related to their holdings. It also provides a uniform interface for the scientific and industrial communities to access the comprehensive microbial resource information.
However, with the development of modern sequencing technology and the aggravation of the world’s energy and environmental crisis, microbial resources are explored even more deeply. In response to this growing importance, the integration of microbial resources for massive data sharing is underway. In the era of Big Data, we should create new data standards and deploy new technologies of integration and visualization to integrate microbiological resources data and in order to achieve a more sufficient exploitation of these resources.
In an age in which microbial data is diversifying rapidly and exploding in volume, the CODATA Task Group on Advancing Informatics for Microbiology (TG-AIM) focuses on microbial data integration standards and technology to promote the application of Big Data approaches in the field of microorganisms. It does this by supporting the development of an effective information environment that promotes and sustains microbial research data activities and by exploring the value of microbiological data in order to better serve to research and industry.
Activities and Outputs for 2015-2016
- Linked data of microbiology
By applying RDF and other big data technologies, WDCM is creating an integrated data warehouse which contains all related microbiological data including the strain information, ‘omics’ data and reference data. A standard ontology and URIs will be employed in the system which will greatly enhance the accessibility and further crosslink of the data.
- Analyzer of Bio-resource Citation (ABC)
ABC is a software tool which extracts citation information from various resources like papers, patents and genome sequences. ABC mines and analyses the data statistic of strain papers and patents among the registered culture collections all over the world and it will also add a new index about usage classifications for papers and patents. Furthermore, achieving the depth of application analysis on strain papers and patents excavations, increasing strain preservation information data, developing the reports of strain researching history and discovering the situation of strain application analysis are also included in the ABC plan for the next 2 years. In addition, an ‘importance degree’ model will be established to develop strain research and application. 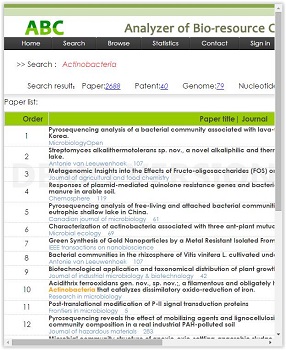
- Information system and analysis tools for Whole Genome Sequence
Taxonomic culture collection catalogs have an inherent bias that may limit discovery. Modern sequence or barcode based taxonomy, based upon core genome characteristics, may overlook pan-genome characteristics in bacteria and archaea, and lineage specific or dispensable chromosomes in fungi. These secondary genome components may carry valuable traits but have distribution that does not always coincide with core genome phylogeny.
At the time of writing in 2014, over 450 strains have been completely sequenced at the JGI and data analysis is ongoing. Among these are 219 genetic loci where a single mutant strain is being sequenced and over 70 loci for which multiple strains or alleles are being studied. WDCM will cooperate with genomic scientists to develop data standards and information system for storing and analysis the whole genome sequence and find a way to integrate the genomic information with the microbial resources information and hence to construct a Big Data model for microbiology.
- Annual Conference and training course
WDCM will hold the Annual International Conference and training course to promote the international bio-information communication and share the world data information technology in next two years.